|
The first step of most image interpretation problems including microscopic
image processing is feature extraction. Feature vectors represent or summarize a
given image. In many problems, image classification is then performed using the
feature vectors instead of actual pixel values. Ordinary feature extraction
methods are developed for regular images and they may not work optimally for
microscopic images. Furthermore, microscopic images are huge in size, which
requires the feature extraction process to be computationally efficient in order
to rely on supercomputers or computer clusters in the future.
One of the feature extraction methods that we are going to try will be based on
the covariance method, which was recently introduced in [15,16,17] and
successfully used in image texture classification and object recognition.
Covariance method features are extensively used in texture classification and
recognition in image and video. Texture classification techniques can be used on
microscopic images to classify them into groups and recognize specific types of
diseases.
The covariance matrix of an image region is defined as follows:
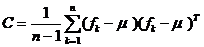 , (1)
, (1)
where fk is the feature vector, containing various parameters of the
k-th pixel such as color component values, wavelet coefficients and gradients.
µ is the mean vector
The computation complexity of the covariance matrix is relatively large ( where
is the size of the image). Computational cost becomes especially important when
a large image needs to be scanned at different scales. Due to the large size of
FL images, supercomputers are used for processing microscopic images at OSU.
Recently, BILKENT developed a co-difference method that can replace the
covariance method in feature extraction. In the co-difference method, an
operator based on additions instead of the multiplications in the covariance
method is used. This methodology gives comparable results in regular texture
classification [18]. The new operator is defined as follows:
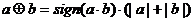 .
(2) .
(2)
This operator is a monoid function, i.e. it satisfies totality, associativity
and identity properties. Therefore it is a semi-group on real numbers. The
operator is basically a summation operation but the sign of the results behaves
like the multiplication operation. As a result, the computational complexity of
microscopic image analysis can be decreased significantly.
Another important issue that needs to be addressed is the choice of entries of
the vector. The obvious choice will be the color pixel values, however, these
may not be descriptive enough. In addition, wavelet or wavelet packet
coefficients may be included in the feature vector. WARWICK has developed a
wavelet transform-domain method, which adaptively selects the best wavelet
features that are optimal in discriminating between different classes of
histology images. The best choice for the feature vector of microscopic images
will be investigated in this WP. It may even be possible to have an adaptive
selection of wavelet coefficients for different parts of an image. Ph.D.
students will work on this problem at BILKENT, WARWICK and OSU. The feature
vectors developed in WP will be used in the 2nd and 3rd WP’s.
|
Work
package no. |
1 |
Start
End
Month |
September
2009
September
2010 |
|
Work
package title |
Microscopic Image Representation and Feature Extraction |
|
Partners Involved |
BILKENT,
OSU, WARWICK, GRECAN |
Objectives:
Develop
computationally efficient feature extraction schemes for microscopic images
using covariance and co-difference matrix methods based on multi-scale
wavelet transform coefficients
Tasks:
-Task
1.1: Find the best feature vector for FL images. Determine which wavelet
scales should be used.
-Task
1.2: Compare the covariance and co-difference methods in H&E images of
FL.
Deliverables:
1) Report on
current methods of feature extraction for microscopic images.
2) Report on
wavelet analysis of microscopic images for best feature selection.
3) Covariance
matrix based feature extraction algorithm.
4)
Co-difference matrix based feature extraction algorithm.
5) Final
report on the scientific work carried out in WP1 during the exchange
programme.
Researchers Involved:
-BILKENT:
Prof. A. Enis Cetin, Alexander Suhre, Kivanc Kose
-OSU: Dr.
Metin N. Gurcan, Olcay Sertel, Jeffrey Prescott
-WARWICK: Dr.
Nasir Rajpoot, Researcher 1, Researcher 2
-GRECAN: Dr. Paulette Herlin, Dr. Myriam Oger
|